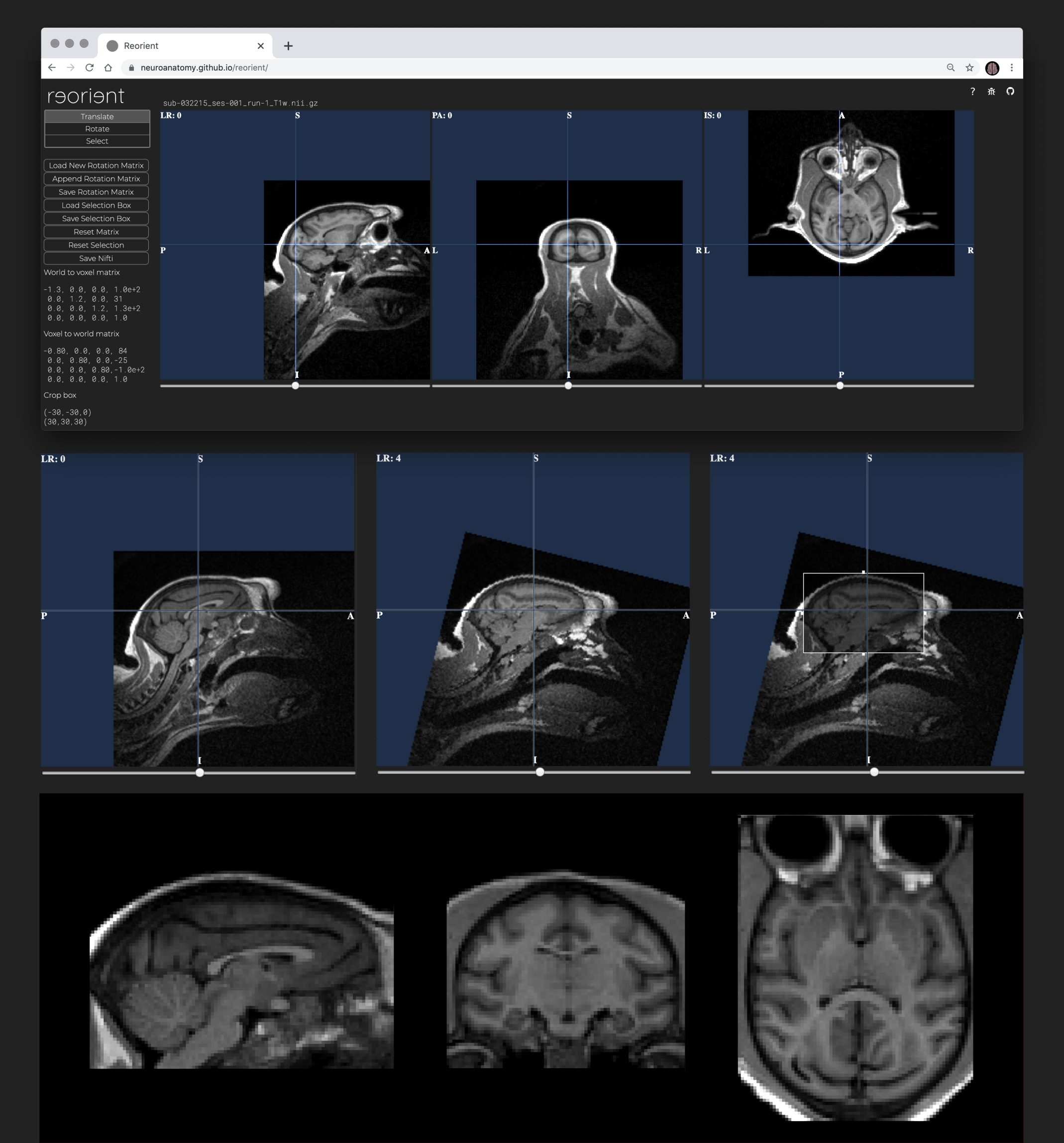
Load & navigate data. To load your data, simply drag and drop your nifti volume (.nii.gz or .nii) onto the upload box. Your MRI will appear in a stereotaxic viewer. Navigate through the slices using the sliders below each viewer.
Translation. Click into the viewer and drag your volume to the desired position. The 3 viewers are synchronised and updated in real-time as are the matrices displayed in the left panel. The translate tool is selected by default when the data has been loaded.
Rotation. Select the Rotate tool in the left panel, and then drag your volume clockwise or counterclockwise into the desired position. The 3 viewers are synchronised and updated in real-time as are the matrices.
Cropping. Click the Select tool in the left panel and drag the 4 boundaries of the selection box to enclose the desired part of the image. Note: The selection box is very small by default. An alert will remind you in case you forgot to adjust it.
Saving. Save the rotation matrix and the selection box together with your reoriented nifti volume for a fully reproducible workflow.
Load new rotation matrix. Load an affine matrix from a text file, and replace the original_matrix in the current MRI by the loaded matrix.
Append rotation matrix. Load an affine matrix from a text file and append it to the one in the current MRI, i.e. change the original matrix into original matrix * loaded matrix.
Reset matrix. Reset the MRI affine matrix to its original value.
Reset selection. Reset the selection box to its default size (small box at center).
Load new MRI file. Simply reload the webpage to start working with a new MRI.
Reproduce the workflow offline. Once you downloaded the rotation matrix and selection box, you will be able to reproduce the reoriented cropped MRI volume using the reorient.py script. It will take the source MRI, the reorient.mat file, and the selection.txt file to produce the reoriented volume, as in python reorient.py input_nifti rotation_file selection_file output_nifti.
How to cite reorient. Heuer, K and Toro, R (2020). Reorient: A Web tool for reorienting and cropping MRI data. Journal of Open Source Software, 5(53), 2670. https://doi.org/10.21105/joss.02670.
🥰 Thank you.