applied & theoretical
The group of Neuroanatomy of the Unit of Human Genetics and Cognitive Functions is interested in the development and evolution of brain anatomy. Our main tools are mathematical modelling, magnetic resonance imaging and human genetics. We develop computational neuroanatomy methods base on our theoretical models which we use to analyse the normal diversity of human brain anatomy, as well as to look for differences associated with neurodevelopmental pathologies. We are particularly interested in Autism Spectrum Disorders. The group of Neuroanatomy is lead by Roberto Toro.
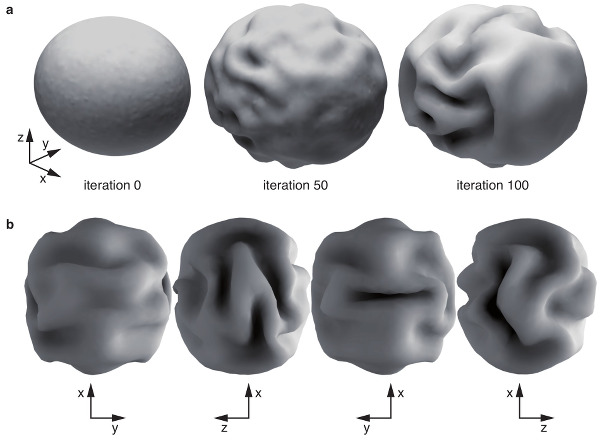
Computational model of cortical folding.
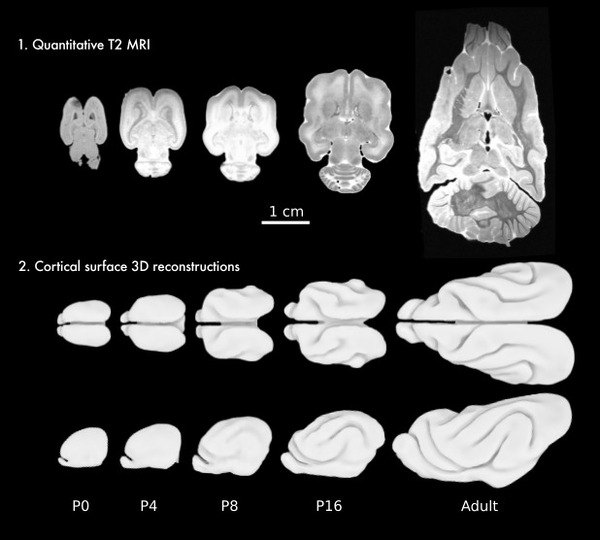
Development of brain folding in ferrets.
Morphogenesis of neocortical organisation: theoretical and animal models
Neocortical folding is one of the most intriguing characteristics of the mammalian brain. Their pattern is very distinctive for different species, and there seems to be a remarkable relationship between convolutions and the architectonic and functional regionalization of the cerebral cortex. Yet the mechanisms behind the development of convolutions and their association with the cortical regionalization are poorly understood. We aim at building models of neocortical morphogenesis based on its geometry, mechanical properties and the growth. Our computer simulations suggest that folding is a natural consequence of cortical growth, and our models are able to reproduce several aspects of folding development, such as the relationship between cortical surface and brain volume among mammals, the period of compensation in the degree of convolution observed in gyrencephalic brains and the dependence of the degree of convolution on cortical thickness. We are interested in studying the effect of early cortical regionalisation on the development of folding, the development of primary, secondary and tertiary convolution, and the relationship between folding, structural and functional connectivity.
[bioRxiv]
Ferrets are exceptional animal models to study brain development, and can provide important information for the interpretation of human neuroimaging. They possess a subdivided subventricular zone, and a rich cytoarchitectonic arealisation, with ~60 cortical areas, similar to a cat. Ferrets are among the smallest mammals to develop the characteristic folds of complex brains. Unlike humans, whose brains start to fold during the 3rd trimester of gestation, and have at birth a complexity similar to that of adults, ferrets are born with a smooth brain and start to develop folds after 1 week. The brain of ferret kits is very immature, and the first weeks after birth provide us with a privileged window to study the development of the relationship between folding and neocortical cytoarchitecture, connectivity and function. We are currently looking at retracing the expansion of the ferret cerebral cortex from birth up to the 10th postnatal week using ultra-high field MRI and high-throughput 3D histology.
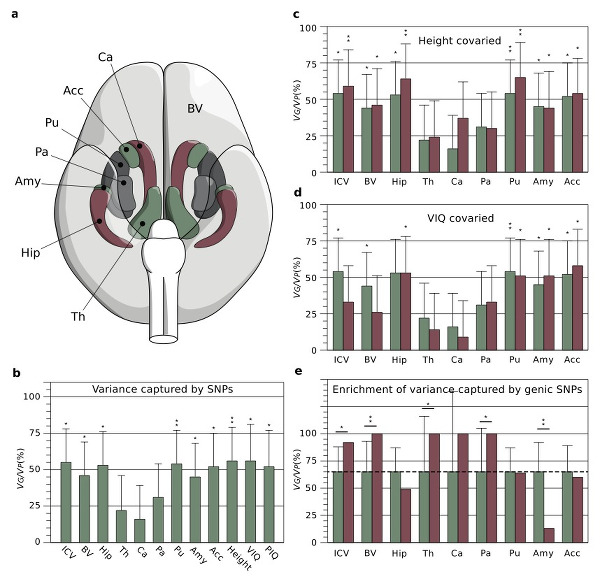
Human neuroanatomical diversity is captured by
thousands of common genetic polymorphisms.
Genomic architecture of normal and pathological human neuroanatomical diversity
Human brain anatomy is strikingly diverse and highly inheritable: genetic factors may explain up to 80% of its variability. Genome-wide scans looking for associations between genomic polymorphism and neuroanatomical variability identify, however, just a few statistically significant signals and account for <1% of the variance. We are currently investigating the genomic architecture of neuroanatomical diversity using a genetic model where the encoding of traits is strongly polygenic. We have been able in this way to show that up to 54% of the heritability of many brain structures is captured by large numbers of genetic polymorphisms of small-effect spread throughout the genome, especially within genes and close regulatory regions. The genetic bases of neuroanatomical diversity appear to be relatively independent of those of body size (height), but shared with those of verbal intelligence scores. The study of this genomic architecture should help us better understand brain evolution and disease.
[Mol Psych] [bioRxiv] [pdf][supp]
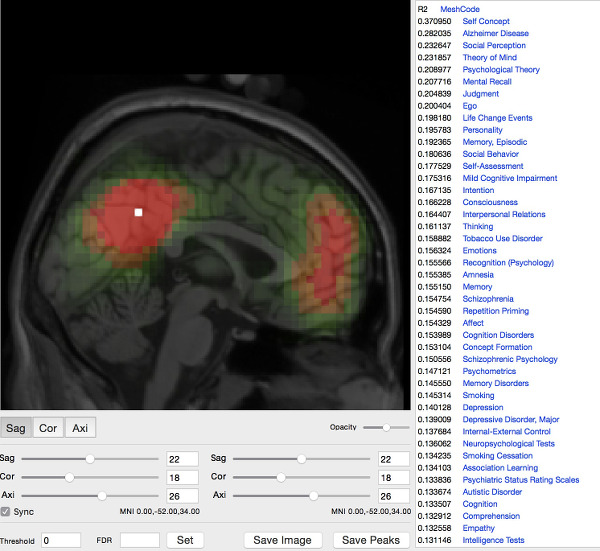
CoactivationMap is a meta-analytic map of
human functional coactivation networks.
Tools for computational Neuroanatomy
We implement and developed data analysis pipelines to process large neuroimaging-genetics datasets, and participate in the analysis of several very large international consortia. We have developed online and offline interactive tools to examine the results of many different pre-processing steps (segmentation of many regions, cortical surfaces, etc.) for hundreds or thousands of subjects.
We are also developing tools to visualise and analyse large high-resolution datasets such as those produced in 3D histology. Histological processing introduces various types of artefacts, distortions, rips, folds, etc., which had to be corrected manually. Once this first preprocessing step is finished, different automatic algorithms can be used for tissue segmentation, classification or cell-density estimation. However, the precise segmentation necessary to build a detailed brain atlas still done manually. We are developing an open, online, neuroinformatic framework to help the scientific community collaborate in the tasks that still require manual intervention, such as quality control, data annotation, and data edition – a neuroinformatic crowdsourcing framework.
Crowdsourcing has proven extremely successful in projects such as EyeWire, where online users are mapping the complete connectivity of a mouse retina, or Synapse, which allows scientists to share projects and compete to find a solution. We are developing a series of web projects exploring the utilisation of crowdsourcing for human neuroimaging, for example BrainSpell and The Brain Catalogue. We have also developed QCCC, an online crowdsourcing to quality control Freesurfer's automatic segmentation of the corpus callosum of >1,000 subjects from the ABIDE cohort.